The Ouverney Lab
 Understanding Complex Microbial Communities
Understanding Complex Microbial Communities
The focus of our research is to better characterize complex microbial communities by type and function. These communities are made of thousands of different species of bacteria. We are particularly interested in uncultured environmental Bacteria and Archaea closely related to microbes associated with humans. By applying culture-independent molecular techniques, we can identify, quantify, and measure metabolic activity in situ of uncultured microbes at the single cell level. Most microbes associated with the human body play a beneficial role in human health and many can also be found in a range of environmental sites. In our pursue to better understand the role of uncultured microbes, we use a holist approach and interrogate both human-associated and environmental complex microbial communities.
Current Projects
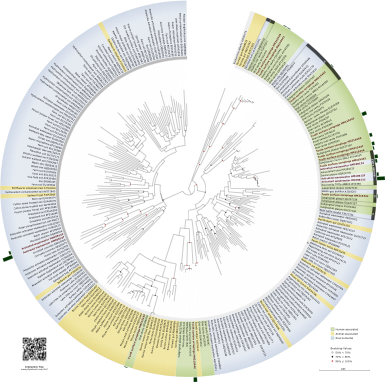
TM7 Bacteria. The uncultured TM7 bacterium phylum (aka Candidatus Saccharibacteria) is associated with the human dental plaque of many individuals. But finding a reliable and consistent oral source of this bacterium is challenging, since humans are constantly cleaning their teeth through brushing, flossing, and mouth washing. Through bioinformatics, the Ouverney lab, in collaboration with the local wastewater treatment plant identified a close relative of the human oral strain in activated sludge in the early 2000s. We also pioneered in using STARFISH (a technique that combines FISH with micro-autoradiography) to measure metabolic activity of TM7 bacteria in situ. Dinis et al 2011. Since then, we have identified novel lineages of this bacterium group, which may play an important role in human health.
Figure 1. FISH micrographs from a confocal microscope depicting bacteria from the uncultured TM7 Phylum (stained in blue with a TM7-specific fluorescent oligonucleotide probe) among many other bacteria from a natural microbial community (stained green with a general DNA dye SYBR-green). Confocal images provide a window to visualize cell morphology and to quantify the prevalence of uncultured microbes such as TM7. Scale bars = 10 µm.
------------------------------------------------------------------------------------
Enrichment of Complex Microbiomes with CRISPR. The human oral microbiome plays a crucial role in human health, and it has been associated with cardiovascular diseases. The advancement of DNA sequencing technologies led to the discovery of many new bacteria associated with dental plaque and a better understanding of its complexity. Nonetheless, studies from around the globe have shown that even NGS is limited in its capacity to survey microbial communities of such abundance and diversity. In fact, 90% of NGS sequences of the human oral microbiome fall within only 5 Bacteria phyla (there are over 70 major Bacteria phyla). We have recently proposed a novel approach that uses CRISPR technology to eliminate the dominant bacterial lineages from our samples while enriching the rare and currently undetectable DNA templates. Even though rare bacteria are found in small proportions (≤ 1% of the total), combined they represent an important group in the homeostasis of the commensal microbiome. We expect this novel approach will reveal new lineages of bacteria associated with humans and eventually a more holistic understanding of their function.
Figure 2. Enrichment of Rare Bacteria in Mixed Natural Communities. CRISPR Cas9 enzyme coupled with bacterial group specific RNA guides are added to
the sample DNA and allowed enough time to digest the target DNA templates of the most
abundant bacterial lineages. By the end of this process, only the undigested DNA
remains. That undigested DNA is expected to be representative of the rare bacteria,
which represent an important conglomerate of the microbiota.
------------------------------------------------------------------------------------
Soil rhizosphere to address global pollution. Though debatable, coffee has been associated with numerous health benefits. Intensive coffee farming practices, however, have been associated with environmental damage, risks to human health, and reductions in biodiversity. In contrast, organic farming has become an increasingly popular alternative, with both environmental and health benefits. Biological species diversity has long been thought to confer benefits in sustainability, with increased diversity providing resistance to stress, disturbance, and changing soil conditions. Bacteria that enhance plant growth, referred to as plant growth-promoting bacteria (PGPB), associated with the coffee rhizosphere can increase crop production by acting as plant growth promoters or supplying plants with nutrients, as demonstrated with isolates such as the phosphate-solubilizing bacteria and nitrogen-fixing bacteria. In 2015 we published a novel report that aimed to characterize and determine the differences in the prokaryotic composition in the soil of Brazilian coffee farms using Next Generation Sequencing. Statistically significant differences were found in the soil rhizosphere microbiome between intensive and organic farms. We were able to identify several groups of bacteria that could be enriched in the soil to enhance coffee production while decreasing the usage of fertilizers and pesticides. These PGPBs have the potential to improve coffee production while diminishing negative environmental impacts at a global scale. Caldwell et al 2025.
Figure 3. Network interactions in Brazilian coffee soil show co-occurrence of rhizobacteria
are dependent on sample site. The network of Operational Taxonomic Units (OTUs) formed
three distinct modules that associated with sample site assigned through LEfSe. The
network contains 199 nodes (circles) and 585 edges (lines). Each node represents an
OTU, and each edge represents a significant (p<0.01) Spearman correlation (rho ≥ 0.6).
The thickness of the edges directly related with the Spearman correlation coefficient.
Node sizes were based on the number of edges connected to the node. Green edges represent
the positive correlation between two nodes. Nodes were respectively colored red (conventional
coffee), blue (transitional coffee), and green (organic coffee), to reflect sample
site assignment of OTU as determined through LEfSe.
Contact Information
Cleber Ouverney
Phone: 408-924-4806
Email: cleber.ouverney@sjsu.edu